Long-interspersed nuclear elements (LINEs) are genetic components found in higher eukaryotes. They are retrotranposons, meaning that they are transcribed into mRNA and then translated into proteins that act as a reverse transcriptase. The reverse transcriptase makes a copy of the LINE DNA which can then be integrated into the genome at a new site. The only active LINE in humans is LINE-1. It has been associated with oncogenesis and Haemophilia A, a diseased caused by insertional mutagenesis.
Current Paper
In this paper published in Cancer Research in Nov. 2018, the researchers set out to explore the function of KDM4B, a histone demethylase that regularly overexpresses in various cancers. It mainly functions by demethylating H3K9me3 in different tumors, resulting in altered gene expression. They carried out a genome-wide analysis on the effects of KDM4B on H3K9me3 distribution in tumor cells. In this study, ABclonal's H3K9me3 antibody was used to target against the protein in various cancer cell lines.
The researchers, Xiang et al., found that H3K9me3 is mainly enriched in LINE-1. And abnormal expression of KDM4B promoted LINE-1 expression and KDM4B depletion reduced expression. When overexpressed, KDM4B "enhanced LINE-1 retrotransposition efficacy, copy number, and associated DNA damage, presumably via the histone demethylase activity of KDM4B."
The findings hint at KDM4B as a regulator for LINE-1. This new role discovered could provide clues to new cancer prevention strategies and therapies.
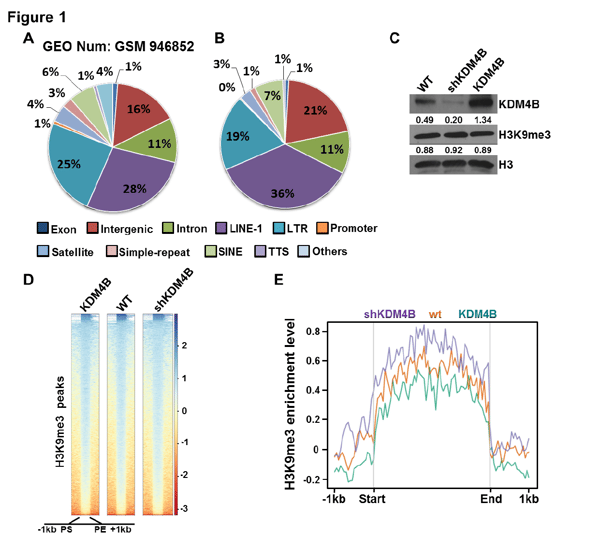 Figure 1. Genome-wide H3K9me3 distribution and the effect of KDM4B on H3K9me3
Figure 1. Genome-wide H3K9me3 distribution and the effect of KDM4B on H3K9me3
A and B, The genomic analysis of distribution of H3K9me3 ChIP-seq peaks with published datasets (GEO number: GSM946852) in MCF7 cells(A) and our H3K9me3 ChIP-seq datasets of MCF7 cells (B), The proportion of each DNA elements that incorporated in H3K9me3 peaks are listed in the pie charts.
C, Immunoblots showed that global H3K9me3 level did not change in MCF7 or MCF-7 derived stable cell lines. H3 was used as internal control. The gray value of the bands was quantified by ImageJ software, the relative values were obtained after normalization by loading control and lablled below the corresponding band.
D, Heatmap display scaled (± 1kb of flanking region) H3K9me3 peaks in MCF7 or MCF-7 derived stable cell lines, intensity scale indicates level of read enrichment.
E, Average profiles showing the H3K9me3 enrichment level at 430 KDM4B-dependent H3K9me3 peaks of the three cell types.
ABclonal's H3K9me3 polyclonal antibody has been validated using WB, IF, IHC, IP, ChIP, and ChIP-seq. Apart from Cancer Research, it has also been featured in journals including Nature Communications, Genome Research, and more.
Reference
Xiang, Y., Yan, K., Zheng, Q., Ke, H., Cheng, J., Xiong, W., ... & Wang, P. (2018). Histone demethylase KDM4B promotes DNA damage by activating long interspersed nuclear element-1. Cancer Research. canres-1310.