One of the most important, most studied, yet still unresolved question in life science is “how can DNA (which unfolds to 2-3 meters in length) fit in the nuclei of eukaryotic cells (which is only a few microns in diameter) and regulate genome functions in an orderly fashion?”
Recently, researchers showed that the nuclear matrix protein HNRNPU could play a universal role in 3D genome.
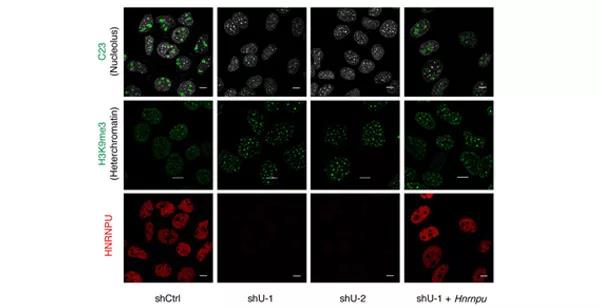
In the experiment, ABclonal epigenetic antibody that targets H3K9me3 was used as a critical reagent.
Experimental Details
The team of researchers established a cell model that attempts to explain "the molecular machinery underlying hierarchically three-dimensional (3D) chromatin architectures." They then systematically examined the regulatory function of nuclear matrix protein HNRNPU (Heterogeneous Nuclear Ribonucleoprotein U) on 3D genome using methods including Hi-C (High-throughput/resolution chromosome conformation capture), DamID (DNA adenine methyltransferase identification), ChIP-Seq, RNA-Seq, and other techniques, including electron microscopy.
The researchers found that when the expression of HNRNPU was down-regulated, the interaction between chromatin and the Lamina-associated domains (LADs) changed dramatically. The result was that 16.6% of the genome transformed from non-LADs to LADs, and the electron microscopy showed a redistribution of chromatin to the nuclear edge. At the same time, 7.5% of the genomes were found to undergo compartment-type transition. Finally, 46% of the TAD borderline intensity and 58% of the chromatin loop intensity decreased. ChIP-Seq experiments showed that HNRNPU mainly binds to the active chromatin region, and 80% of the binding sites overlap with CTCF or RAD21. The study systematically analyzed the global regulatory function of HNRNPU on the chromatin structure and suggested that the nuclear matrix could play a universal role in the 3D genome. In addition, it provided a new basis for revealing the molecular mechanism of chromatin formation and maintenance in higher structure.