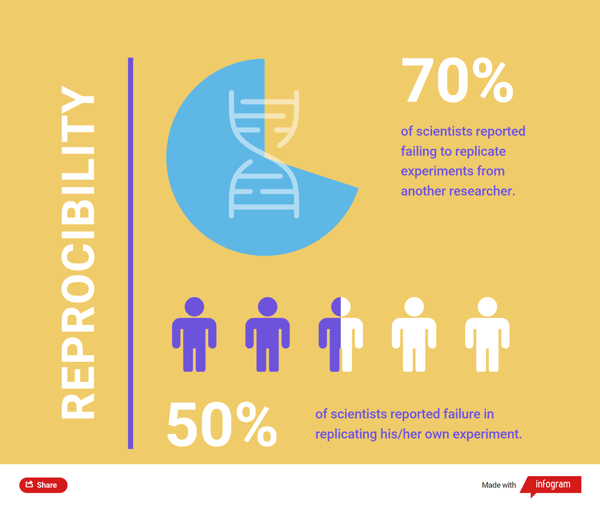
The scientific community operates on a self-correcting model that relies on repetition and replication. However, according to a 2016 survey by Nature, more than 70% reported to have failed to replicate experiments from another scientist, more than 50% reported failure in replicating his/her own experiment. Out of the 1,576 scientists surveyed, 906 were from biology or medicine disciplines.
To further exacerbate the problem, a 2015 study in PLoS Biology estimated the potential cost of irreproducible preclinical research in the United States at $28 billion per year. The same study also cited poor-quality reagents as the largest contributor to irreproducibility. Antibodies, as one of the most commonly used reagents in biology research, seem like a good target to tackle in order to solve the problem at hand.
Antibody specificity is the most important factor when it comes to antibody quality. In 2016, researchers from the International Working Group for Antibody Validation (IWGAV) published a paper that recommends methods to validate antibody specificity, one of which is through genetic strategies – by testing cell or tissue samples with the target protein depleted and compare the results to samples with the protein expressed at normal levels (Uhlen et al., 2016).
How to Deplete Target Proteins?
Target protein(s) can be depleted using gene knockout. Generally speaking, gene knockout is used to investigate the function of a gene (of which the sequence is known). It refers to the gene editing technologies that inactivate (knock out) a gene through replacement or disruption of DNA. Once the gene(s) encoding the target protein(s) is inactivated or deleted, a protein-depleted sample can be obtained.
3 Gene Editing Tools for Gene Knockout
Traditionally, homologous recombination was the main method for gene knockout. However, this method is time consuming and inefficient. Currently, there are three prominent genome-editing tools that are more efficient and more precise compared to traditional methods: zinc-finger nucleases (ZFN), transcription activator like effector nucleases (TALEN), and clustered regularly interspaced short palindromic repeats (CRISPR).
ZFN and TALEN are both chimeric nucleases composed of a programmable, sequence-specific DNA-binding domain (zinc finger DNA-binding domain/TAL effector DNA-binding domain) and a nuclease. Once the DNA-sequence-specific binding domain binds to the target gene, the nuclease cleaves the DNA and causes a double strand break (DSB). The cells utilize their repair mechanism through non-homologous end joining (NHEJ) which introduces mutations that destroy functionality of the gene.
CRISPR on the other hand, relies on Cas9, an RNA-guided DNA endonuclease to target and edit specific gene sequences in situ. This method is simpler, more efficient, and more cost-effective than ZFN and TALEN.
In an effort to help mitigate the waste associated with irreproducibility, ABclonal is validating our antibodies using CRISPR/Cas9, the leading technology in gene-editing. Contact us to find out more about our 100+ KO-validated antibodies and how they may help improve the reliability and reproducibility of your research.
Additionally, if you're curious about other aspects of antibodies in research, take a look at these four other blogs for further reading.