Large-scale sequencing projects have great potential to provide a wealth of knowledge to scientists and the public. Perhaps the most celebrated project of this nature is the Human Genome Project (HGP) which was completed in 2003. For many, the multi-billion endeavor was considered a “moonshot” for biology, but with its successful completion (99% of the euchromatic genome sequenced with 99.99% accuracy) came the launch of many other large-scale sequencing projects such as the Cancer Genome Atlas (2005) or more recently, the Earth Bio-genome Project (2018). The introduction of large-scale quantitative methods, such as next-generation sequencing, have also made these projects feasible.

Recently, an international team of scientists from the United States, China, and Canada collaborated on an ENCODE project to survey RNA-binding proteins on the chromatin regions of the human genome. The researchers used chromatin immunoprecipitation and RNA-sequencing methods to identify binding regions and to quantify the effects of RBPs on transcription output. The work was completed as an ENCODE project, which was a large-scale sequencing project that started in 2003 as the logical step to follow the Human Genome Project. While HGP mapped the nucleotide base pairs making up human DNA, ENCODE was built to identify the functional elements in the human genome.
The goal of ENCODE is to build a comprehensive parts list of functional elements in the human genome, including elements that act at the protein and RNA levels, and regulatory elements that control cells and circumstances in which a gene is active. - ENCODE
The long-term value of such exploratory-based projects is hard to imagine or measure. Any large-scale sequencing project, even the Human Genome Project, is a major investment requiring global collaboration. However, once completed, the application for such data is potentially limitless. For example, HPG ultimately led to numerous medical advances, starting with the diagnosis and treatment of cancers.
Lately, RNA-binding proteins have garnered renewed attention from scientists as recent proteome-wide studies revealed that there is over double the amount of RBPs than previously thought. For a long time, RNA-binding proteins were mainly studied for their roles in the RNA life cycle (in metabolism events such as synthesis, folding, or degradation), but in a recent review article, scientists concluded that RBPs may be one of the most abundant proteins expressed in the cell. The same analysis also presented a census containing more than 1500 RBPs. In light of these findings, researchers have become curious about other functions RBPs might have besides their roles in RNA metabolism.
In the ENCODE study published in Cell, the team of scientists wanted to explore the potential for RBP involvement in transcriptional activities due to the fact that many RNA-processing events are tightly coupled with transcription. Evidence of the integration of RBPs in transcription would help explain how transcriptional and post-transcriptional machineries are intertwined.
To start their discovery-driven project, the scientists needed to first establish the presence of RBPs on chromatin. To narrow down the list of RBPs, the team started with proteins localized in the nuclei of two human cell lines, HepG2 and K562. Fifty-eight and forty-five RBPs from the KepG2 and K562 lines, respectively, were studied due to the availability of antibodies. Although custom antibody projects are possible, the survey still creates a starting point for adding RBPs to the ENCODE database.
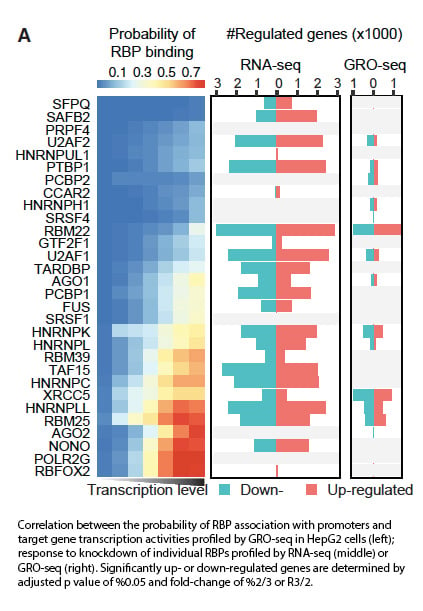
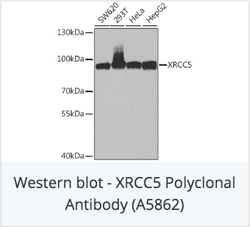
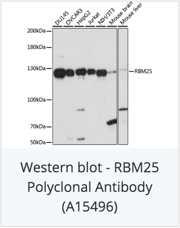
With ChiP sequencing, the scientists found that RNA-binding proteins were in fact predominantly (~60%) localized on active chromatin regions. In particular, all chromatin-associated RBPs had preference for binding to active gene promoters, hinting at their potential involvement in transcription as previously hypothesized. Knowckdown of individual RBPs showed that six RBPs (RBM22, XRCC5, RBM25, HNRNPK, HNRNPLL, and U2AF1) had considerable direct impacts on output, as measured by RNA-seq. These RBPs induced differential expression of over 500 genes after knockdown.
Check out catalog antibodies for RNA-binding proteins:
If you're looking for other helpful reads regarding proteins, be sure to check out our other related blogs here.
.jpg)