The arguably most fun thing about science is when your supervisor tells you to just do Experiment X to test hypothesis, but then they kind of forget to tell you how complicated the techniques are to perform that experiment, not to mention all the optimization you would need to do. I personally have never done a chromatin immunoprecipitation (ChIP), and since I wasn’t in genomics, the most sequencing I ever did was setting up quick reactions for the core facility to tell me that my gene constructs were correctly built. ChIP does sound rather simple when explained in class, but when you read up on the protocols,1 there are some limitations to what ChIP can do, especially given the large amount of starting material you need for the typical experiment. Luckily, in recent years, scientists have started to use an alternative technique called Cleavage Under Targets and Tagmentation, or CUT&Tag, which ABclonal is pleased to support through our antibody reagents.
The arguably most fun thing about science is when your supervisor tells you to just do Experiment X to test hypothesis, but then they kind of forget to tell you how complicated the techniques are to perform that experiment, not to mention all the optimization you would need to do. I personally have never done a chromatin immunoprecipitation (ChIP), and since I wasn’t in genomics, the most sequencing I ever did was setting up quick reactions for the core facility to tell me that my gene constructs were correctly built. ChIP does sound rather simple when explained in class, but when you read up on the protocols,1 there are some limitations to what ChIP can do, especially given the large amount of starting material you need for the typical experiment. Luckily, in recent years, scientists have started to use an alternative technique called Cleavage Under Targets and Tagmentation, or CUT&Tag, which ABclonal is pleased to support through our antibody reagents.
The History of CUT&Tag
The original CUT&Tag methodology was devised and reported by Dr. Steven Henikoff’s group at the Fred Hutchinson Cancer Research Center. 2 The goal was to develop an efficient method that could map chromatin using as little starting material as a single cell. Not sacrificing quality for cost, the method appears to be robust, allowing scientists to generate high-resolution epigenetic maps with short turnaround and comparatively lower expense. The Henikoff group had previously developed CUT&RUN, another method that was motivated by the researchers’ gripes with ChIP, including the fact that ChIP protocols usually run the course of several days.
In contrast to ChIP, CUT&Tag and CUT&RUN don’t require cells to be cracked open and their DNA and chromatin to be broken into pieces. Instead, cells stay intact, making the approach applicable to single-cell analyses, including projects like the Human Cell Atlas.
Henikoff Lab postdoctoral fellow Dr. Hatice Kaya-Okur — another lead scientist on the team — worked with Henikoff and Ahmad to improve CUT&Tag so that it compresses the DNA-processing from two steps into one. This makes it possible to produce sequencing-ready DNA snippets in a single day. In contrast, ChIP and CUT&RUN take a few days to get to that step. The team also automated most of the process to speed it even further.
Imagine a streamlined protocol that includes the building of your DNA library in the same step from just a few cells, or even one cell, that can also be automated. And the bonus is that CUT&Tag will give you comparable, even superior, data compared to ChIP or traditional ChIP-sequencing (ChIP-seq)!
How Does CUT&Tag Work?
To overcome the limitations of ChIP, ChIP-seq, and their earlier innovation of CUT&RUN, Henikoff’s scientists used a transposase called Tn5 that they describe as “hyperactive,” which is also fused to Protein A and is loaded with sequencing adapters normally used in next-generation sequencing platforms. 2 The basic premise is shown in this infographic.
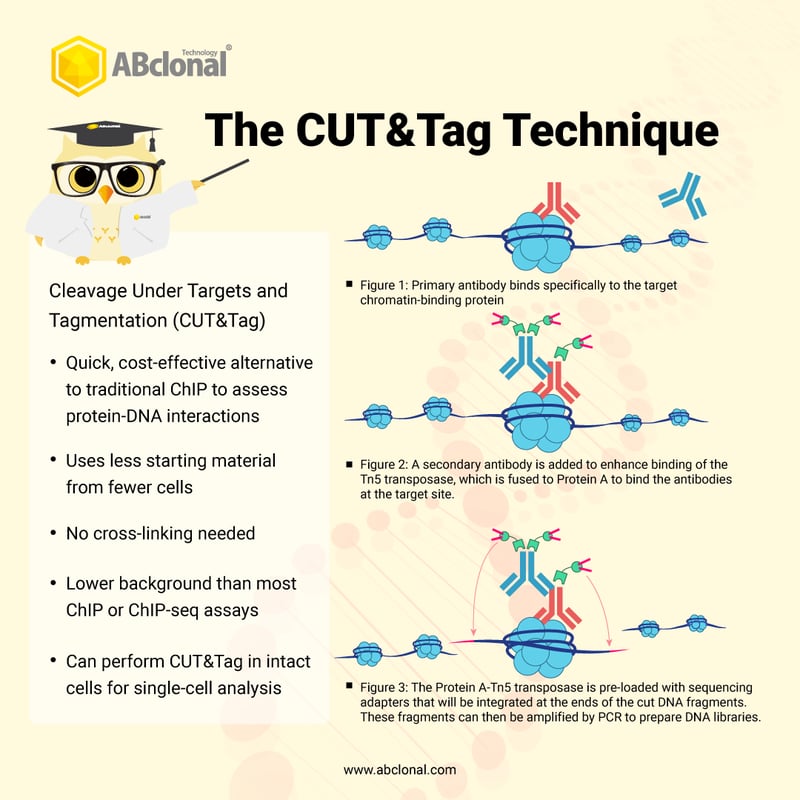
In the first step, the cells (which can be worked with in the absence of fixing agents like formaldehyde) are permeabilized to allow the application of a primary antibody that targets the chromatin-interacting protein. After a wash step, a second antibody is added to bind to the primary, similar to a Western blot or ELISA. The purpose of both antibodies is to leverage the Protein A fused to the Tn5 transposase and ensure targeted binding to the desired site. After the wash step, the transposase is activated by addition of magnesium (Mg2+), which allows the transposase to cleave the DNA sites around the target binding site while also integrating the pre-loaded sequencing adapters. In this way, the DNA library setup is performed in a single step prior to PCR amplification. Automation and the use of solid supports allows for enrichment of the target sites of interest. 2
Advantages of CUT&Tag
CUT&Tag was invented to alleviate the complications the authors experienced with traditional ChIP techniques. With CUT&Tag, genomes and protein-DNA interactions can be analyzed using very few cells, and the authors even described some single-cell experiments because the cells are intact and do not experience the crosslinking reactions required by ChIP. Also, the CUT&Tag does not require sonication, so is far less invasive and violent to reduce sample loss and to maintain protein epitopes and complexes.
 One can imagine that it would be difficult to isolate too many cells of certain types, including stem cells, so CUT&Tag is beneficial to researchers who have a limited supply of starting material, and it is adaptable for single-cell analysis. 3 Since CUT&Tag focuses on the chromatin proximal to the target binding site, fewer sequencing reads are required to generate quality data with lower background. The superior signal-to-noise ratio and consistency of the CUT&Tag method persists even with novel applications, like this study that established a native G4 map. 4 With its ease of use and lower cost, it is no wonder more scientists are moving to CUT&Tag for their studies.
One can imagine that it would be difficult to isolate too many cells of certain types, including stem cells, so CUT&Tag is beneficial to researchers who have a limited supply of starting material, and it is adaptable for single-cell analysis. 3 Since CUT&Tag focuses on the chromatin proximal to the target binding site, fewer sequencing reads are required to generate quality data with lower background. The superior signal-to-noise ratio and consistency of the CUT&Tag method persists even with novel applications, like this study that established a native G4 map. 4 With its ease of use and lower cost, it is no wonder more scientists are moving to CUT&Tag for their studies.
How ABclonal Can Help
With the rise in popularity of the CUT&Tag technique, ABclonal has begun to validate our ChIP-ready antibodies for use in CUT&Tag applications. Please see below for a list of some of our initially CUT&Tag validated antibodies. More antibodies will be validated in the coming months, so please contact us if you have a target in mind that you would like to explore in your next study!
|
Cat. No. |
Product |
|
Acetyl-Histone H3-K27 Rabbit mAb |
|
|
[KO Validated] CDK9 Rabbit mAb |
|
|
Histone H3 Rabbit mAb |
|
|
[KO Validated] c-Myc Rabbit mAb |
|
|
CTCF Rabbit mAb |
References
- Nelson JD, Denisenko O, and Bomsztyk K (2006) “Protocol for the fast chromatin immunoprecipitation (ChIP) method.” Nature Protocols 1:179-185.
- Kaya-Okur et al. (2019) “CUT&Tag for efficient epigenomic profiling of small samples and single cells.” Nature Communications 10:1930 (Epub).
- Bartosovic M, Kabbe M & Castelo-Branco G (2021) “Single-cell CUT&Tag profiles histon modifications and transcription factors in complex tissues.” Nature Biotechnology 39:825-835.
- Lyu et al. (2022) “Genome-wide mapping of G-quadruplex structures with CUT&Tag.” Nucleic Acids Research 50(3):e13 (Epub).