My first experience in a basic research laboratory was a structural biology project, in which we were attempting to solve the structure of a nervous system protein known as myelin basic protein (MBP). As a rookie undergraduate scientist at the University of California, I had great mentors who taught me everything, from how to purify recombinant proteins from bacteria to doing library work to understand what had been done before so I could build upon it. I also learned how to use an electron microscope (EM) to gather structural data. MBP was an interesting challenge as it had multiple isoforms due to alternative splicing, and generally behaved like a random coil. 1 The major function of MBP is to take advantage of its highly positive charge to compact myelin in higher organisms, with research over the years suggesting it may have some capacity to form alpha helices, although atomic-resolution structures have not yet been reported. MBP has also been reported as a biomarker in autoimmune diseases such as multiple sclerosis. 1
My first experience in a basic research laboratory was a structural biology project, in which we were attempting to solve the structure of a nervous system protein known as myelin basic protein (MBP). As a rookie undergraduate scientist at the University of California, I had great mentors who taught me everything, from how to purify recombinant proteins from bacteria to doing library work to understand what had been done before so I could build upon it. I also learned how to use an electron microscope (EM) to gather structural data. MBP was an interesting challenge as it had multiple isoforms due to alternative splicing, and generally behaved like a random coil. 1 The major function of MBP is to take advantage of its highly positive charge to compact myelin in higher organisms, with research over the years suggesting it may have some capacity to form alpha helices, although atomic-resolution structures have not yet been reported. MBP has also been reported as a biomarker in autoimmune diseases such as multiple sclerosis. 1
While I ended up in a different field, the lessons I learned from structural biology helped me better appreciate how the conformation of a protein contributes to our understanding of its function in the cell and organism.
The Central Dogma: Not As Simple As it Sounds!
All biology students have heard of the central dogma, which states that the genetic code (stored in DNA) is transcribed into messenger RNA and then translated into proteins by ribosomes. What isn’t emphasized is how many decades of research it took to arrive at the principle we take for granted now. Ever since Christian Anfinsen’s landmark discovery that won the 1972 Nobel Prize in Chemistry for connecting the protein’s amino acid sequence to its biological activity, we have had a greater appreciation for how structure leads to function. Although it is more complicated than just letting thermodynamics work, a properly folded protein is critical to normal cellular functions and organismal health.
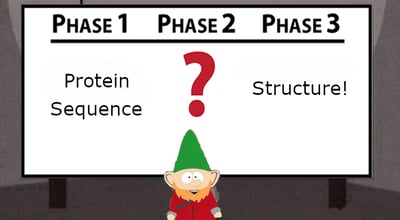
The easy part of the project, now that the Human Genome Project was completed and technology has advanced, is determining the amino acid sequence from the candidate gene. The hard part, as many have discovered, is figuring out how that sequence folds into the final, biologically functional conformation. Assuming the protein can be expressed and purified in large enough quantities to be processed into a crystal, the next step is to perform procedures such as x-ray crystallography, nuclear magnetic resonance (NMR), mass spectrometry, cryo-EM, and other techniques to piece together enough data to generate a high-resolution structure. 2 As you can imagine, many of these methodologies are resource-heavy, labor-intensive, and time-consuming, so like many folks who would prefer to do literally anything else, structural biologists are always looking for a shortcut.
The Holy Grail: Predicting Protein Structures?
With the accelerated growth of computers and computing power, researchers began to develop computational modeling methods to bridge the gap between spotty experimental data and a more refined structure. 2, 3 The structural biologist’s dream is to be able to feed an amino acid sequence into a machine, turn the crank, and spit out a structure. In a bid for further progress as well as to stroke some egos, research groups have participated in the Critical Assessment of Structure Prediction (CASP) competition to display their newest prediction algorithms and strategies. The protein structure prediction strategies are split into template-based modeling (TBM) that utilize reference structures deposited into the Protein Data Bank (PDB), or template-free modeling (FM) that do not rely on existing structures. 2, 3 These methods rely on machine learning and probabilistic models, with known physics and chemistry principles thrown in, to fit the structure. 3
The proficiency of these models has improved over the past three decades since CASP started, and in 2021, two major publications were published describing new platforms that are freely accessible and predict structures with unprecedented accuracy. 4, 5 These accomplishments were recognized by Science Magazine as the breakthrough of 2021, being much more adept at computing structures in a fraction of the time needed to do a crystallography experiment, and allowing fellow scientists to continue contributing to the broad knowledge base without barriers.
The Future of Protein Folding
 More work is required before we achieve the Star Trek scientific utopia where you can just wave a tricorder over a sample and know everything about it instantly, but the ability to train algorithms on experimental data, even if that data is incomplete, is very encouraging for the future of structural biology. Even with the advancement of protein prediction software, traditional experiments are still required to not only provide reference data on which to build a model, but also to confirm the structure in biological context. 2 Limitations still exist for even these breakthrough programs, as they may not be as capable of solving multi-domain protein structures or accommodate protein complex structure predictions of interactions between different proteins. 3 Although the goal is to be able to achieve structural predictions without doing experiments, the laboratory is still indispensable.
More work is required before we achieve the Star Trek scientific utopia where you can just wave a tricorder over a sample and know everything about it instantly, but the ability to train algorithms on experimental data, even if that data is incomplete, is very encouraging for the future of structural biology. Even with the advancement of protein prediction software, traditional experiments are still required to not only provide reference data on which to build a model, but also to confirm the structure in biological context. 2 Limitations still exist for even these breakthrough programs, as they may not be as capable of solving multi-domain protein structures or accommodate protein complex structure predictions of interactions between different proteins. 3 Although the goal is to be able to achieve structural predictions without doing experiments, the laboratory is still indispensable.
Despite the imperfections, the freely accessible software and ease of use provide plenty of avenues for advancement going forward. I hope that this encourages the scientific community to continue to be collaborative, to keep as much of science open access as possible, and given the nature of these experimental methods, to implement coding and statistical analysis into their research. Science is better when we can all share our stories, and this breakthrough has made it easier for us to exchange knowledge for the betterment of humanity.
BioChat: Native Proteins!
Lots of research is based on recombinant proteins these days, but there is still a market and a use for proteins purified directly from the source species. Check out this episode of BioChat where I hang out with Josiah Carney of ProNique Scientific to talk about how native proteins still have advantages and are still useful today!
Please check out our BioChat page to learn how to subscribe (and of course please rate and review us on Apple Podcasts!) and check out our latest episode by clicking the player above. Thanks for listening!
References
- Raasakka & Kursula (2020) “Flexible Players within the Sheaths: The Intrinsically Disordered Proteins of Myelin in Health and Disease.” Cells 9(2):470 (Epub; Correction 2022).
- Seffernick & Lindert (2020) “Hybrid methods for combined experimental and computational determination of protein structure.” J Chem Phys 153(24):240901 (Epub).
- Pearce & Zhang (2021) “Toward the solution of the protein structure prediction problem.” J Biol Chem 297(1):100870 (Epub).
- Tunyasuvunakool et al. (2021) “Highly accurate protein structure prediction for the human proteome.” Nature 596:590-596.
- Baek et al. (2021) “Accurate prediction of protein structures and interactions using a three-track neural network.” Science 373(6557):871-876.